|

|
|
In this section of my research, I have collaborated to various
artificial life studies, which focused on the level of large
populations of individuals and examined their collective evolution.
Although there are no "morphogenetic engineering" concerns per se,
individuals are still internally sophisticated, as they can
contain a long genomic sequence, an intricate genetic program, or a
set of generative (rewrite) rules giving rise to complex morphologies.
|
|
|
A spatially explicit model of endogenous speciation in the absence
of environmental constraints

|
 |
A commonly held view in evolutionary biology is that speciation,
i.e., the emergence of genetically distinct and reproductively
incompatible subpopulations, is driven by the external environment.
Guy Hoelzer, Rich Drewes and myself have developed a
spatially explicit model of a biological population to study the
emergence of spatial and temporal patterns of genetic diversity in
the absence of such constraints.
|
|
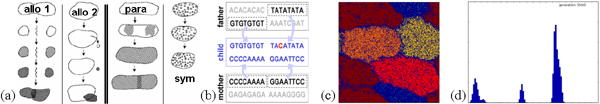
Speciation is usually thought to be either caused by geographical
boundaries ("allopatric" speciation from mountains, rivers, islands,
etc.; Fig. a, left) or pre-existing environmental
inhomogeneities ("parapatric" or "sympatric" speciation from
unequal climate or resource distribution; Fig. a, right).
We propose a 2-D cellular automata model showing that an
initially homogeneous population might spontaneously segment into
different species through sheer isolation by distance,
without the need for natural divisions.
Each location on the lattice contains several individuals that
follow simple rules of reproduction, mutation and migratory
dispersal. At each generation, they randomly move within a
certain distance, mate with other individuals in their
neighborhood and produce offspring, which can also mutate with a
small probability.
Their diploid genomes comprise two pairs of 1000-base
chromosomes assorted independently during mating (Fig. b).
Due to an "outbreeding depression" effect, offspring viability
is also bound to decrease with the genetic distance between
parents.
|
|
|
|
Our simulations show that in a certain domain of parameters a
phase transition revealing spontaneous segmentation and
clustering occurs in the network. This inherent tendency toward
spatial self-organization is analogous to Turing pattern formation
on the population scale (Fig. c).
To explore this pattern of population diversity we plot a
frequency histogram of the genetic difference between pairs of
randomly chosen individuals, or "mismatch distribution"
(Fig. d). While a well-mixed version of the model
exhibits a relatively stable and unimodal distribution, a drifting
multimodal distribution emerges when the dispersal of individuals is
limited to short distances. The long-term persistence of such
diverging subpopulations is the essence of biological speciation.
|
|
Publication
|
|
|
A model of spatial population dynamics combining L-systems, genetic
expression, biologically inspired mutations, and open-ended evolution

|
 |
This part of José David Fernández's PhD, which I co-directed with
his main advisor Francisco Vico,
examines the formation, evolution, and diversity dynamics of
a community of virtual plants through a new
individual-centered model at multiple scales:
genetic, developmental, and physiological. It is
an original attempt to combine development, evolution, and
population dynamics (from multi-agent interactions) into one
comprehensive, yet simple model.
|
|
Understanding the dynamics of biodiversity has
become an important line of research in theoretical ecology and,
in particular, conservation biology. However, studying the evolution
of ecological communities under traditional modeling approaches
based on differential calculus requires species' characteristics to be
predefined, which limits the generality of the results.
An alternative
but less standardized methodology relies on intensive computer
simulation of evolving communities made of simple, explicitly
described individuals. Compared to analytical frameworks,
two major properties are distinctive of the agent-based computational approach: emergence
and adaptation. New properties at higher scales, which were not explicitly included in the model,
appear in the system as the individuals interact. Simultaneously,
the community can also evolve and adapt to new environmental conditions, therefore broadening
the scope of possible experiments and testable hypotheses.
In this work, we focus on an especially puzzling aspect of evolution: diversification. We simulate a
virtual community of plants to study the emergence and dynamics of genomic and phenotypic variation
during evolution. To model plants, we use rewriting systems, a family of formal methods widely
investigated in theoretical computer science and capable of encoding and generating complex structures.
In this formal framework, parts of an initial object, generally strings of symbols, are iteratively
replaced by other parts, generally longer string segments, following rewrite rules.
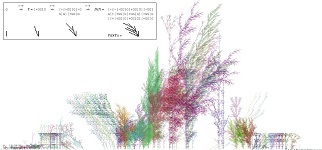
We observe that our
simulated plants evolve increasingly elaborate canopies, which are
capable of intercepting ever greater amounts of light. Generated
morphologies vary from the simplest one-branch structure of
promoter plants to a complex arborization of several hundred
thousand branches in highly evolved variants. On the population
scale, the heterogeneous spatial structuration of the plant community
at each generation depends solely on the evolution of its component plants.
Using this virtual data, the morphologies and the dynamics
of diversity production were analyzed by various statistical methods,
based on genotypic and phenotypic distance metrics. The results
demonstrate that diversity can spontaneously emerge in a community
of mutually interacting individuals under the influence of specific
environmental conditions.
|
|
Publication
|
|
|
Exploring a new class of cellular automata capable of longterm phenotypic
dynamics, including a high level of variance and behavioral diversity.

|
 |
In this original work by my MSc student David Medernach, co-supervised by Taras Kowaliw, we study open-ended evolution by analyzing
heterogeneous cellular automata (HetCA). The model involves cells that have
an "age", "decay", and "quiescence", a transition function based on genetic
programming, and transfer between adjacent cells. These changes convert
a CA system into a new kind of "ecosystem", where different genomes
compete for existence.
|
|
A major motivation for work in artificial life is to create
open-ended evolution, or systems in which novel artifacts are
continuously produced. The best known example is Ray's Tierra
world, where there is competition between replicating computer
programs in a virtual machine. Later, another evolutionary
system by Adami, called Avida, became popular and was extended
in many directions. High-level systems exist as well, where agents
execute complex predefined functions, such as eating or fighting
in Yaeger's Polyworld.
In all these cases, there is no particular specified goal:
interesting and sometimes unexpected phenomena emerge
from the interaction of individuals and their environment.
An important insight into the majority of such successful
systems is that they involve ecosystemic interactions.
|
|
|
|
Our new artificial life model is based on a discrete dynamical systems
framework. We have extended classical 2D cellular automata (CA), our chief
modification being the allowance for heterogeneous transition functions.
The cumulative effect of these features is to create an evolving ecosystem
of competing cell colonies.
To evaluate our new model and demonstrate its open-endedness,
we define a measure of phenotypic diversity on the space of cellular automata.
Our results show that HetCA is capable of supporting long-term dynamical
behaviour and phenotypic changes, not readily achieved
in control homogeneous CA, such as the Game of Life. In particular, we exhibit
examples of the strategies discovered by our system, which are characterized
by the emergence of competitive behaviour.
|
|
References
Ray (1991) An approach to the synthesis of life. ALife II.
Yaeger (1993) Polyworld. ALife III. SFI Series.
Adami & Brown (1994) Avida. ALife IV. MIT Press.
|
Publications
|
|